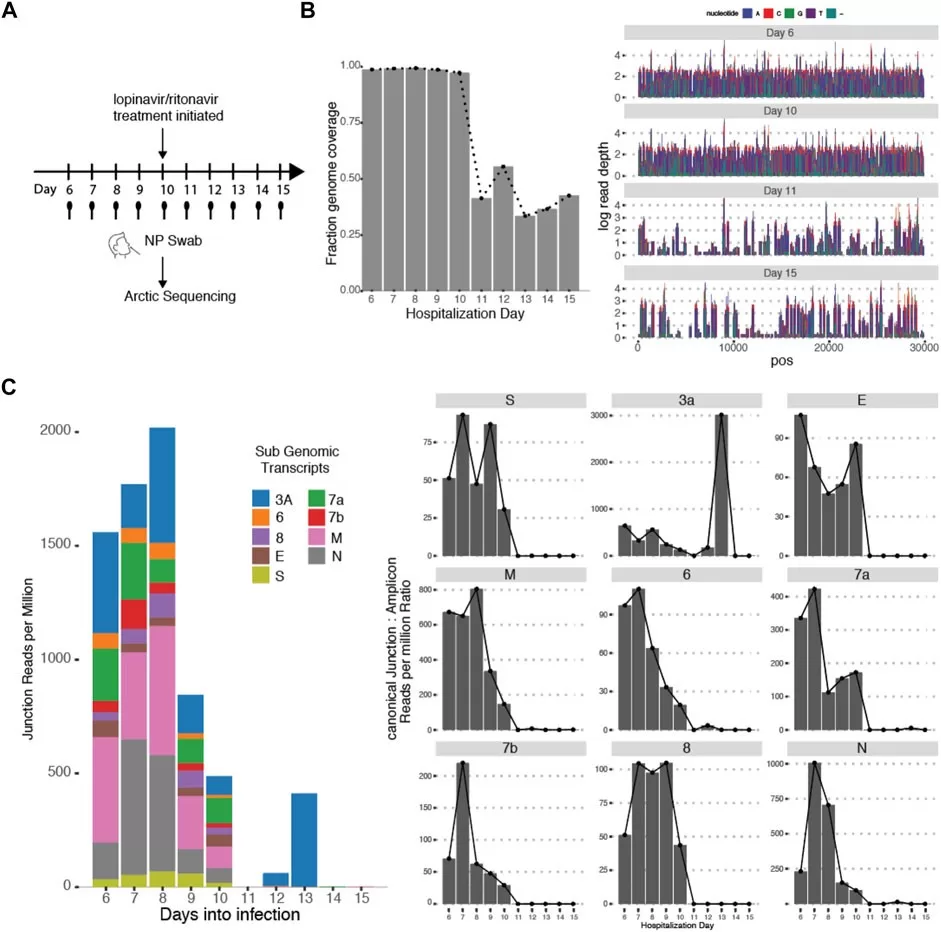
SARS-CoV-2 genomic surveillance plays a crucial role in understanding the evolution of the virus and guiding public health responses during the pandemic. As COVID-19 continues to impact communities, the ability to analyze genetic sequences of the virus is essential in tracking variants and informing vaccine development. Traditional methods relied heavily on nucleic acid amplification tests (NAATs), but as their use decreased following the end of the public health emergency, innovative approaches were needed. Our study demonstrates how community health initiatives, such as utilizing rapid antigen tests (RATs) for genomic sequencing, can maintain robust surveillance efforts. This strategy not only enhances the volume of available genomic data but also fosters community engagement in public health initiatives.
The continuous monitoring of SARS-CoV-2 is integral to public health, especially in the evolving landscape of COVID-19 variants. Genomic analysis, through alternative testing methods, opens a new avenue for effective disease management as traditional testing methods become less accessible. This initiative highlights the importance of integrating technologies such as COVID-19 genomic sequencing and rapid antigen tests in community frameworks. By leveraging community resources, we can improve our understanding of the virus and enhance public health protocols. Such approaches not only support accurate tracking of transmission patterns but also facilitate proactive community health responses.
The Role of SARS-CoV-2 Genomic Surveillance in Controlling COVID-19
SARS-CoV-2 genomic surveillance plays a crucial role in controlling the COVID-19 pandemic by monitoring the virus’s mutations and variants. This specialized surveillance provides critical data that informs public health responses, enabling authorities to identify and combat emerging variants of concern effectively. As the pandemic evolves, understanding the genomic landscape of SARS-CoV-2 is essential for tailoring vaccination strategies and public health mitigations. Through robust genomic sequencing, health officials can pinpoint changes in the virus that may influence transmissibility or vaccine effectiveness.
The integration of SARS-CoV-2 genomic surveillance into public health frameworks enhances overall community health initiatives. By collaborating with local health departments, the genomic data obtained can help in formulating strategies that directly address the needs of the population. In this context, continuous genomic monitoring facilitates timely interventions, particularly in areas identified as hotspots for COVID-19 transmission. This proactive approach can significantly reduce infection rates and save lives, demonstrating the value of genomic surveillance as not just a reactive measure, but as a fundamental component of effective public health strategy.
Community Engagement in Genomic Surveillance
Community involvement has proven instrumental in sustaining genomic surveillance efforts, especially following the shift from traditional laboratory-based testing to more accessible options like rapid antigen tests (RATs). Engaging local populations in submitting RAT samples can yield a treasure trove of genomic data while simultaneously fostering a sense of communal responsibility towards public health. The partnership between health authorities and local entities such as libraries creates a supportive environment that encourages widespread participation. This collaborative model enhances the reach and effectiveness of genomic surveillance, ensuring that even after the public health emergency, monitoring can continue effectively.
Moreover, the success of integrating RATs into the genomic surveillance framework illustrates the potential of community-based initiatives. By removing barriers to access and providing easy instructions for recording and submitting positive results, communities are empowered to take an active role in health surveillance processes. This model not only improves data collection but also builds a stronger relationship between public health agencies and the community, enhancing trust and adherence to public health guidelines. The key takeaway is that community engagement can significantly fortify our response to public health challenges, with genomic surveillance being a pivotal aspect.
Advancements in Nucleic Acid Amplification Techniques
Advancements in nucleic acid amplification tests (NAATs) have revolutionized the capacity for researchers to conduct genomic analyses. These tests, particularly valuable for their sensitivity and specificity, have enabled the rapid identification of SARS-CoV-2 variants in various populations. As the pandemic evolved, the application of enhanced amplification techniques allowed for more accurate viral genome sequencing even in the changed landscape of testing availability. Despite the challenges faced post-public health emergency declaration, the integration of innovative methodologies in genomic sequencing remains pivotal in our ongoing response to COVID-19.
Utilizing refined techniques in nucleic acid extraction and amplification is essential not only for SARS-CoV-2 but could also extend to the surveillance of other respiratory viruses. The methods applied in this study demonstrate a merging of technology with public health initiatives, contributing to a robust model for future genomic surveillance efforts. Continuous improvement of these techniques facilitates large-scale testing and provides public health officials with the necessary tools to preemptively address potential outbreaks, reinforcing the critical nature of evolving diagnostic methodologies.
The Impact of Public Health Responses on Genomic Surveillance
Public health responses have a profound impact on the effectiveness of genomic surveillance strategies. The quality and quantity of data derived from genomic sequencing depend significantly on the robustness of testing infrastructures and the responsiveness of health authorities to emerging threats. In Wisconsin, the end of the COVID-19 public health emergency on May 11, 2023, led to reduced reliance on nucleic acid amplification tests. However, the initiative to incorporate rapid antigen tests into the genomic mix has successfully bridged that gap, underscoring how adaptive public health responses can maintain surveillance capabilities even in challenging contexts.
This adaptability in public health responses not only aids in genomic monitoring but also positions communities to be more resilient against future viral outbreaks. By integrating lessons learned from the COVID-19 pandemic, public health officials can design more effective frameworks for genomic surveillance. This involves not only focusing on current methodologies but also planning for scalable implementation that incorporates community feedback and engagement, fostering a collaborative public approach to health management.
Community Health Initiatives for Sustained Surveillance
Community health initiatives are essential for ensuring the sustainability of genomic surveillance, particularly in light of the reduced availability of traditional testing methods post-COVID-19 emergency. By organizing accessible programs for community members to participate in, local health departments can secure a continuous stream of data essential for understanding the viral landscape. The model employed in Dane County, Wisconsin, highlights the importance of creating frameworks that facilitate community participation, thereby enhancing the overall public health response to infectious diseases.
Effective community health initiatives can also drive awareness around the importance of genomic sequencing and surveillance. Educating the public about the significance of submitting rapid antigen tests strengthens the community’s role in public health strategy. The collaboration with entities such as library systems adds an educational aspect that emphasizes the shared responsibility of protecting community health. This educational approach fosters an environment of partnership that empowers citizens to participate actively in health initiatives, ultimately leading to more effective genomic surveillance outcomes.
Incorporating Rapid Antigen Tests into Genomic Monitoring
The incorporation of rapid antigen tests (RATs) into genomic monitoring represents a crucial pivot point for public health efforts in managing COVID-19. These tests provide a low-barrier option for individuals to quickly assess their COVID-19 status, allowing for timely sampling and subsequent genomic analysis. By actively involving communities in this process, health authorities can accumulate valuable genomic data while also facilitating immediate responses to potential outbreaks. The versatility of RATs also helps maintain surveillance capabilities in environments where traditional testing has become less accessible.
Furthermore, leveraging RATs for genomic sequencing allows for a more comprehensive understanding of viral spread and mutation patterns. As seen in Dane County, Wisconsin, these tests provide a practical and efficient method for sustaining genomic surveillance efforts. Analyzing the viral genome sequenced from RAT samples can yield insights into emerging strains that may evade current public health measures or vaccines. Consequently, rapid antigen tests become not only a tool for individual diagnosis but also a powerful asset for public health monitoring and strategic planning.
Engagement with Local Health Departments for Effective Surveillance
Engaging with local health departments is a critical element in establishing effective SARS-CoV-2 genomic surveillance programs. Such collaborations ensure that data collected from genomic sequencing aligns with broader public health objectives, enhancing the impact of surveillance efforts on community health outcomes. The partnership between academic researchers and local health agencies facilitates access to necessary resources and expertise, creating a synergistic approach to public health monitoring.
Moreover, these collaborations enable continuous feedback loops that drive improvements in sampling techniques, data collection, and analysis methods. By sharing results with local health departments, researchers can help inform response strategies that address the needs of the community effectively. This dynamic relationship can lead to developing best practices that can be replicated in other regions, promoting a cohesive approach toward genomic surveillance and public health readiness at various levels.
The Future of COVID-19 Genomic Surveillance
The future of COVID-19 genomic surveillance depends on the integration of innovative testing methods and community-oriented strategies. With the lessons learned from reliance on traditional nucleic acid amplification tests, there is a pressing need to adopt flexible and adaptive surveillance frameworks that include rapid antigen tests and other accessible diagnostic tools. By cultivating a robust network of community engagement, local health departments can sustain genomic monitoring initiatives well into the post-pandemic landscape.
Adapting genomic surveillance for the future involves not only technological advancements but also a commitment to continuous public health education. Ensuring that the public understands the importance of participating in genomic surveillance by submitting RATs cultivates a proactive health mindset. As the landscape of infectious diseases continues to evolve, the resilience built through effective community efforts and innovative surveillance techniques will be vital in preventing and managing future outbreaks.
Frequently Asked Questions
What is SARS-CoV-2 genomic surveillance and why is it important?
SARS-CoV-2 genomic surveillance is the ongoing monitoring of the genetic variations of the virus that causes COVID-19. It is crucial for identifying new variants, tracking their transmission patterns, and informing public health responses to effectively combat outbreaks.
How does COVID-19 genomic sequencing work within SARS-CoV-2 genomic surveillance?
COVID-19 genomic sequencing involves analyzing the genetic material of the virus from collected samples, such as those from rapid antigen tests (RATs) or nucleic acid amplification tests (NAATs). This process helps identify specific variants and their prevalence in communities, guiding public health efforts.
What role do rapid antigen tests (RATs) play in SARS-CoV-2 genomic surveillance?
Rapid antigen tests (RATs) provide an accessible source for SARS-CoV-2 samples in genomic surveillance. By collecting and sequencing data from these tests, researchers can effectively monitor variants circulating in the community, especially following the decline in availability of NAATs.
How have public health responses been enhanced by SARS-CoV-2 genomic surveillance?
SARS-CoV-2 genomic surveillance has significantly bolstered public health responses by supplying crucial data on viral variants, enabling timely interventions and the adjustment of vaccination strategies, thus promoting better community health initiatives.
What are some community health initiatives associated with SARS-CoV-2 genomic surveillance?
Community health initiatives linked to SARS-CoV-2 genomic surveillance include the distribution of rapid antigen tests (RATs), public engagement in sample collection, and collaboration with local health departments and libraries to enhance data collection efforts in tracking the virus.
Why are nucleic acid amplification tests (NAATs) significant for SARS-CoV-2 genomic surveillance?
Nucleic acid amplification tests (NAATs) are significant for SARS-CoV-2 genomic surveillance because they are highly sensitive and reliable for detecting the virus. Although their usage decreased post-public health emergency, NAATs initially provided a foundation for genomic data collection that informed responses to the pandemic.
What were the results of SARS-CoV-2 genomic surveillance using community-distributed RATs in Dane County?
In Dane County, SARS-CoV-2 genomic surveillance using community-distributed RATs yielded 128 successful genome sequences from 227 samples, demonstrating a practical and effective approach to maintaining genomic tracking of the virus in local populations.
How does community engagement impact SARS-CoV-2 genomic surveillance efforts?
Community engagement is vital for the success of SARS-CoV-2 genomic surveillance. By actively involving the public in testing and reporting, researchers can gather important data, fostering a collaborative effort to monitor and respond to COVID-19 and other potential respiratory viruses.
| Key Point | Details |
|---|---|
| Background | SARS-CoV-2 genomic surveillance initially depended on leftover diagnostic samples, primarily from nucleic acid amplification tests (NAATs), which became less accessible post-health emergency. |
| Collaboration | The study involved collaboration with local public health agencies and community systems in Dane County to obtain samples from rapid antigen tests (RATs). |
| Sample Size and Sequencing | Out of 227 RAT samples collected between August 2023 and February 2024, 127 sequences were successfully generated with adequate genome coverage. |
| Correlation Observed | Findings suggested a correlation between lower cycle threshold values in RATs and successful sequencing outcomes. |
| Community Engagement | High levels of community participation in submitting RATs were noted, indicating readiness for public health involvement. |
| Future Directions | The study implies RATs can effectively supplement existing genomic surveillance and could be adapted for other respiratory viruses. |
Summary
SARS-CoV-2 genomic surveillance is essential for guiding public health responses to current and future outbreaks. This study demonstrated the feasibility of using rapid antigen tests collected from the community to continue effective genomic monitoring. Through robust community engagement and innovative sample collection strategies, researchers successfully sequenced viral genomes, reinforcing the importance of collaborative efforts in public health. Adaptations of this model may enhance the surveillance of other infectious diseases, showcasing a proactive approach to public health challenges.
The content provided on this blog (e.g., symptom descriptions, health tips, or general advice) is for informational purposes only and is not a substitute for professional medical advice, diagnosis, or treatment. Always seek the guidance of your physician or other qualified healthcare provider with any questions you may have regarding a medical condition. Never disregard professional medical advice or delay seeking it because of something you have read on this website. If you believe you may have a medical emergency, call your doctor or emergency services immediately. Reliance on any information provided by this blog is solely at your own risk.