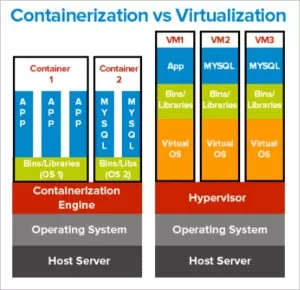
Software containerization in public health has emerged as a transformative strategy to streamline complex processes like genomic surveillance and bioinformatics analysis. By integrating next-generation sequencing (NGS) workflows into containerized environments, public health laboratories can ensure higher reproducibility and efficiency in handling genomic data. This approach not only simplifies the deployment of bioinformatics tools but also enables rapid response capabilities in disease outbreaks. With the rise in infectious diseases and the global impact of the COVID-19 pandemic, the relevance of containerization for genomic surveillance has never been more critical. Embracing these advanced technologies positions public health efforts at the forefront of innovative solutions to manage infectious disease challenges.
In the realm of public health, utilizing container technology to package and deploy software solutions represents a significant advancement in managing bioinformatics tasks. Termed as container orchestration, this method supports the continual evolution of genomics and next-generation sequencing applications. The implementation of standardized software environments facilitates the rapid adoption of bioinformatic workflows, crucial during health crises like pandemics. Moreover, these containerized tools play a vital role in enhancing genomic data analysis, offering scalability and portability across varying public health infrastructures. By harnessing such innovative solutions, public health bioinformatics can significantly improve its analytical capabilities and response times.
The Role of Software Containerization in Public Health Bioinformatics
Software containerization has revolutionized public health bioinformatics by streamlining the management of complex software dependencies required for next-generation sequencing (NGS) analyses. Containers encapsulate an application and its environment, ensuring that bioinformatic tools function consistently across various laboratory setups. By providing a standardized platform, containerization enhances the reproducibility and reliability of genomic workflows—a critical aspect for public health initiatives, particularly during outbreaks of infectious diseases.
Additionally, the adoption of software containerization enables laboratories to rapidly deploy bioinformatic workflows without worrying about system compatibility issues or conflicts between software versions. This agility is especially important in public health settings where genomic surveillance is essential for tracking pathogen evolution and emergence. Consequently, containerized bioinformatics tools not only save time and resources but also bolster public health response efforts by ensuring that effective and accurate genomic analysis can be performed in a timely manner.
Frequently Asked Questions
What are the benefits of software containerization in public health bioinformatics?
Software containerization in public health bioinformatics offers several key benefits, including enhanced reproducibility of next-generation sequencing (NGS) workflows, improved isolation to prevent version conflicts, and greater portability of bioinformatic applications across various computing environments. These advantages streamline the deployment of genomic surveillance tools, making it easier for laboratories to adopt and maintain complex bioinformatic software.
How does software containerization support genomic surveillance in public health?
Software containerization enhances genomic surveillance in public health by providing consistent and reliable environments for running bioinformatic analysis. This leads to improved accuracy in genomic data processing and analysis, which is crucial for tracking disease outbreaks and variants. Containerized applications like those from the StaPH-B project facilitate rapid deployment and ensure that public health laboratories can access the latest tools and protocols needed for effective surveillance.
What role does next-generation sequencing (NGS) workflow play in public health with software containerization?
Next-generation sequencing (NGS) workflows benefit significantly from software containerization in public health as it allows for easy integration of multiple bioinformatic tools while maintaining version control and dependency management. This approach ensures that NGS results are reproducible and reliable across different public health laboratories, essential for effective disease monitoring and response.
Can software containerization improve the efficiency of public health laboratories in managing bioinformatic tools?
Yes, software containerization greatly improves the efficiency of public health laboratories by simplifying the management of bioinformatic tools. Containerized software environments reduce the need for extensive configuration efforts, allowing laboratories to focus more on data analysis than on software maintenance. This efficiency is particularly important in time-sensitive scenarios, such as during disease outbreaks.
What is the StaPH-B project and its significance in bioinformatics containerization for public health?
The StaPH-B project is a significant community-driven initiative that provides a repository of containerized bioinformatic tools tailored for public health applications. During the COVID-19 pandemic, it played a vital role in enhancing genomic surveillance by offering standardized, reproducible workflows for NGS analysis. This project exemplifies how containerization can facilitate collaboration and resource sharing in confronting public health challenges.
How do software containers ensure reproducibility in public health genomic workflows?
Software containers ensure reproducibility in public health genomic workflows by encapsulating all necessary software and dependencies into a controlled environment. This isolation means that researchers can run the same analyses repeatedly without inconsistencies arising from differing software versions or configurations, thereby enhancing the credibility of genomic surveillance efforts.
What challenges does software containerization address for next-generation sequencing in public health?
Software containerization addresses several challenges in next-generation sequencing (NGS) for public health, including managing complex software dependencies, avoiding version conflicts, and facilitating easy deployment across different laboratory infrastructures. By providing standardized and reproducible workflows, containerization helps public health labs maintain high-quality genomic analyses.
What is the importance of bioinformatics containerization during health emergencies like the COVID-19 pandemic?
During health emergencies like the COVID-19 pandemic, bioinformatics containerization is crucial as it allows for rapid development and deployment of genomic analysis tools. This agility ensures that public health systems can quickly adapt to new data and emerging variants, providing timely information essential for managing public health responses.
| Key Points | Details |
|---|---|
| Advantages of Software Containerization | 1. **Reproducibility**: Enhanced control over software versions and dependencies. 2. **Isolation**: Prevents version conflicts and maintains data integrity. 3. **Portability**: Facilitates easy distribution and scalability of bioinformatic tools. |
| Role in COVID-19 Pandemic | The StaPH-B docker project played a crucial role in genomic surveillance, enabling reliable workflows and extensive usage of containerized tools during the COVID-19 pandemic. |
| Impact on Workflow Management | Software containerization simplifies installation and reduces maintenance issues, allowing laboratories to save time and resources while adopting next-generation sequencing workflows. |
Summary
Software Containerization in Public Health has greatly revolutionized genomic workflows, particularly within the context of infectious disease surveillance. By encapsulating bioinformatic software and its dependencies, it provides a reliable, reproducible, and portable framework that aids clinical and public health laboratories in their analysis tasks. The initiatives during the COVID-19 pandemic, including the StaPH-B docker project, exemplify how containerization can enhance efficiency and standardization in public health. This advancement not only improves the management of genomic data but also paves the way for future innovations within the field.
The content provided on this blog (e.g., symptom descriptions, health tips, or general advice) is for informational purposes only and is not a substitute for professional medical advice, diagnosis, or treatment. Always seek the guidance of your physician or other qualified healthcare provider with any questions you may have regarding a medical condition. Never disregard professional medical advice or delay seeking it because of something you have read on this website. If you believe you may have a medical emergency, call your doctor or emergency services immediately. Reliance on any information provided by this blog is solely at your own risk.