Decentralized Sequencing Listeria represents a groundbreaking shift in the approach to genomic surveillance of Listeria monocytogenes in Australia. Between 2016 and 2023, the implementation of decentralized sequencing has dramatically improved turnaround times for genomic data, critical for effective public health investigations monitoring foodborne infections. This proactive strategy not only streamlined the process from sample collection to surveillance reporting but also empowered various jurisdictions to manage their sequencing capabilities. As a result, Australia’s ability to utilize whole-genome sequencing (WGS) for Listeria has enhanced, providing significant insights into outbreak clusters and the overall epidemiology of foodborne illnesses. Through these advancements, the nation exemplifies how decentralized initiatives can strengthen public health frameworks and improve responses to potential listeriosis outbreaks.
The evolution of genomic surveillance mechanisms regarding Listeria leads to essential changes in how health authorities track and respond to bacterial infections. Alternative terms like decentralized genomic analysis of Listeria, local sequencing methods, or independent surveillance programs signify an emerging trend in managing public health data effectively. This progressive transition allows jurisdictions to flexibly adapt their sequencing operations while ensuring data integrity and enhancing regional response capabilities. In this context, the collaboration among various health sectors benefits from the efficiencies gained through whole-genome sequencing, contributing to a comprehensive understanding of Listeria monocytogenes pathology. Ultimately, these advancements position Australia at the forefront of genomic methodologies for addressing critical food safety concerns.
Understanding Listeria monocytogenes and its Impact on Public Health
Listeria monocytogenes is a pathogenic bacterium responsible for listeriosis, an infection that poses significant public health risks, particularly in vulnerable populations such as pregnant women, neonates, and individuals with weakened immune systems. With a relatively low incidence rate, the high case-fatality ratio associated with this foodborne pathogen emphasizes the importance of effective surveillance systems. By leveraging whole-genome sequencing (WGS), public health authorities can more accurately identify outbreaks and trace their origins, leading to more effective prevention strategies and health interventions.
The nuances of public health investigations surrounding Listeria monocytogenes demand a comprehensive approach, where genomic surveillance becomes an essential tool. By integrating detailed genomic data with clinical and epidemiological information, health officials can better understand the pathogenesis of listeriosis and develop targeted responses. In Australia, this is particularly evident in the advancements facilitated by decentralized sequencing, wherein local jurisdictions are empowered to analyze samples rapidly, thus enhancing the efficacy of outbreak management and reducing the overall burden of foodborne infections.
The Role of Decentralized Sequencing in Genomic Surveillance
Decentralized sequencing has revolutionized genomic surveillance frameworks across many regions, particularly in Australia. By empowering individual jurisdictions to conduct their own sequencing, the national Listeria monocytogenes genomic surveillance system has seen marked improvements in turnaround times for sample analysis. Prior to this transition, a centralized laboratory model created bottlenecks, leading to prolonged waiting periods for crucial genomic data. The shift to a more distributed approach has optimized sample throughput, as observed by the reduction in median reporting times from 32 days in 2016 to just 26 days by 2023.
Furthermore, the rollout of decentralized sequencing has allowed for more localized insights into listeriosis trends and outbreaks. This model facilitates rapid responses to emerging public health threats, aligning seamlessly with real-time data needs during incidents of foodborne illness outbreaks. As jurisdictions adapt their capabilities, the sharing of genomic information becomes streamlined, creating a robust network of surveillance that not only addresses immediate public health concerns but also lays the groundwork for enhanced collaborative efforts in future investigations.
Whole-Genome Sequencing as a Tool for Outbreak Identification
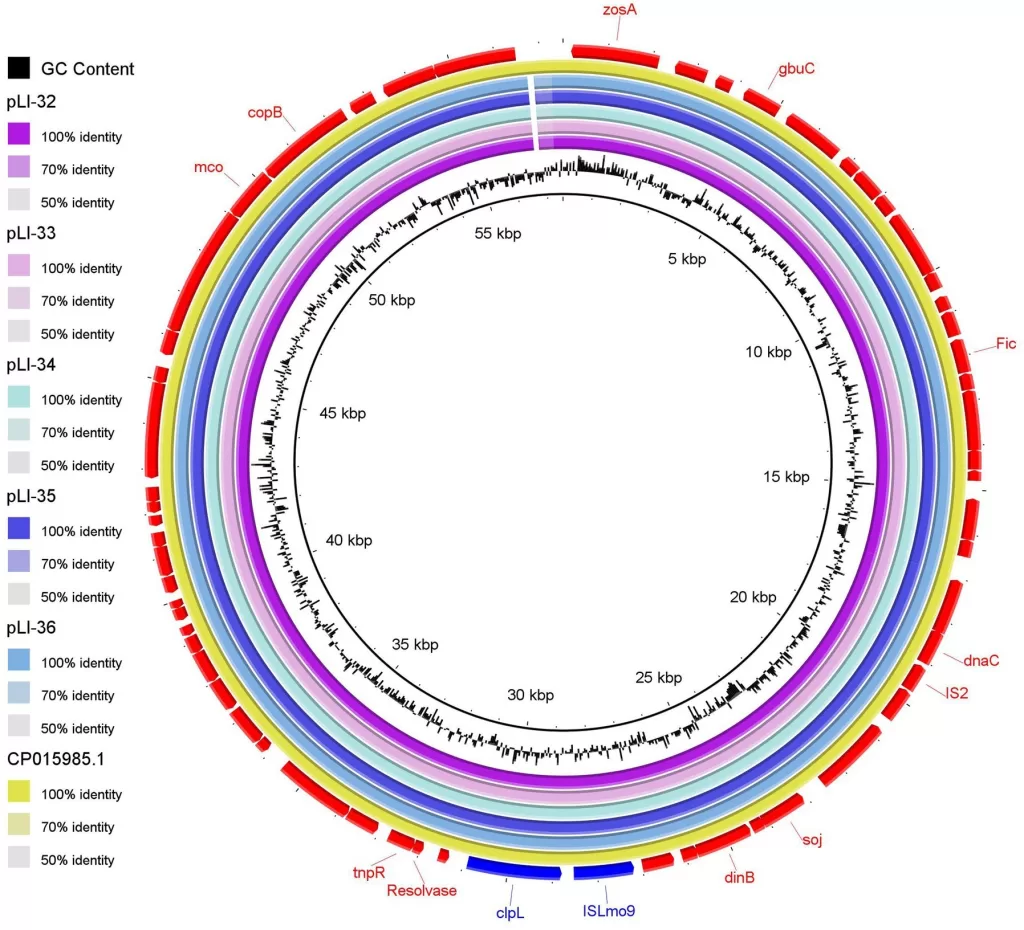
Whole-genome sequencing (WGS) has emerged as a pivotal technology in the identification of Listeria monocytogenes outbreaks due to its high-resolution genetic profiling capabilities. Compared to traditional methods such as pulsed-field gel electrophoresis (PFGE), WGS allows for precise distinction among closely related bacterial strains, which is crucial for tracing sources of infection. The implementation of WGS in genomic surveillance systems enables public health investigators to link cases to potential sources more effectively, thereby preventing further transmissions.
In the context of Australia’s public health landscape, the integration of WGS into the decentralized genomic surveillance framework represents a significant leap forward. It enhances the capacity to monitor listeriosis cases, identify patterns in infections, and correlate environmental samples with human cases. This ability to rapidly assess the genetic makeup of L. monocytogenes not only amplifies the understanding of its epidemiology but also fosters proactive measures in food safety and regulation, significantly mitigating the risks associated with foodborne infections.
Enhancing Food Safety through Improved Surveillance Practices
Enhancements in genomic surveillance practices have profound implications for food safety initiatives. The decentralized sequencing of Listeria monocytogenes facilitates timely identification and response to foodborne outbreaks, effectively preventing widespread illness. Public health authorities can react swiftly to outbreaks, issuing recalls and advisories based on real-time genomic data. This proactive approach not only safeguards public health but also bolsters consumer confidence in food safety protocols.
The lessons learned from Australia’s experience can illuminate pathways for other countries aiming to strengthen their food safety systems. By adopting practices that prioritize rapid genomic surveillance and decentralized sequencing, nations can improve their capacity to manage foodborne infections. Such collaborations across jurisdictions will lead to robust frameworks that enhance public health responses, driven by data-informed decision making and strategic resource allocation.
Statistical Insights into Listeriosis Incidence in Australia
Analyzing statistical data from the National Notifiable Diseases Surveillance System (NNDSS) provides valuable insights into the incidence rates of listeriosis in Australia. During the period from 2016 to 2023, a total of 545 reported cases were closely monitored, and this dataset reveals a unique trend in the outbreak of illnesses associated with Listeria monocytogenes. Notably, the stability in average notifications underscores the effectiveness of ongoing surveillance, particularly given the notable increases in sample testing due to enhanced sequencing capacity.
Furthermore, the significant drop in turnaround times for genetic analysis reflects the positive impact of decentralized methods on public health outcomes. While initial challenges were present as jurisdictions adapted to new sequencing capabilities, the overall results indicate a flourishing system capable of addressing and mitigating threats posed by listeriosis. These statistical insights highlight the importance of evolving public health systems to incorporate modern technologies like whole-genome sequencing, providing a more nuanced understanding of foodborne disease epidemiology.
Challenges in Transitioning to Decentralized Genomic Surveillance
While the potential benefits of decentralized genomic surveillance for Listeria monocytogenes are pronounced, the transition has not been without its challenges. Some jurisdictions experienced initial increases in turnaround times as they navigated the complexities of local sequencing operations. The necessity for clear referral mechanisms became evident, as discrepancies in operational protocols could hinder swift data reporting. Developing a cohesive protocol across jurisdictions is vital, fostering consistency and efficiency as each locale integrates its sequencing capacity.
Another challenge lies in the need for adequate training and resources within local public health laboratories. As jurisdictions take on more responsibility for genomic surveillance, they must build the necessary infrastructure and expertise to analyze and interpret WGS data effectively. Monitoring these transitions and offering support to laboratories will be crucial to ensure that delays do not undermine the overall benefits of decentralized genomic surveillance and that public health responses remain robust.
Future Directions for Genomic Surveillance of Listeria monocytogenes
The future of genomic surveillance for Listeria monocytogenes looks promising, particularly as advancements in technology continue to evolve. Increasing the accessibility of sequencing technologies to public health jurisdictions is likely to yield faster and more reliable data. Moreover, fostering international collaborations can further enhance the understanding of Listeria genomics globally, enabling countries to share insights and tools critical for managing foodborne illnesses.
As Australia has demonstrated, implementing decentralized sequencing can serve as a model for other nations. Emphasizing the importance of real-time data sharing and collaborative outbreak response strategies will ensure that public health officials are equipped to handle the complexities of foodborne infections effectively. With ongoing research and development in WGS technologies, the potential to detect and prevent outbreaks before they escalate has never been more attainable.
The Significance of Timeliness in Public Health Responses
Timeliness is a critical component of effective public health responses, especially in the management of outbreaks caused by Listeria monocytogenes. Rapid laboratory processing and data sharing are essential in curbing the spread of foodborne illnesses. The reduced median time from sample collection to genomic data issuance in Australia is a testament to the advantages that a decentralized sequencing approach can provide. By prioritizing swift responses, public health systems can react effectively, mitigating the risks associated with listeriosis.
Moreover, timely genomic data allows for swift decision-making regarding food safety regulations and outbreak containment measures. The quicker health authorities can pinpoint the source of listeriosis outbreaks, the more effectively they can implement interventions, such as food recalls or public advisories. Ultimately, prioritizing timely genomic surveillance not only protects public health but also enhances the overall infrastructure of food safety systems.
Leveraging Genomic Surveillance for Preventive Health Strategies
The application of genomic surveillance for Listeria monocytogenes significantly contributes to preventive health strategies. By utilizing whole-genome sequencing, public health officials can identify trends, high-risk sources, and potential outbreak strains, allowing them to craft informative and preventative measures ahead of time. This proactive approach shifts the paradigm from reactive interventions to strategic prevention, promoting healthier populations reliant on data-driven decisions.
As the Australian genomic surveillance system adapts to include decentralized sequencing, the overarching narrative continues to focus on prevention and preparedness. Local investigations can lead to impactful food safety reforms and better consumer education, ultimately aiming to reduce the incidence of listeriosis. As countries worldwide look toward improving their public health frameworks, the integration of genomic data into decision-making processes remains paramount in safeguarding against foodborne infections.
Frequently Asked Questions
What is the role of decentralized sequencing in Listeria monocytogenes genomic surveillance in Australia?
Decentralized sequencing plays a crucial role in Listeria monocytogenes genomic surveillance in Australia by enhancing the efficiency and speed of data reporting. This system allows jurisdictions to conduct local sequencing, reducing the turnaround time for genomic data from 32 days in 2016 to just 26 days by 2023, even as sample volumes have doubled.
How has decentralized sequencing affected the public health investigations of Listeria monocytogenes outbreaks in Australia?
The decentralized sequencing of Listeria monocytogenes has significantly bolstered public health investigations in Australia. By enabling faster genomic data collection and analysis, public health officials can more effectively trace outbreak origins and link food and environmental samples to human cases, thereby improving outbreak response strategies.
What improvements in whole-genome sequencing turnaround times have been observed between 2016 and 2023 in the context of Listeria monocytogenes?
Between 2016 and 2023, whole-genome sequencing turnaround times for Listeria monocytogenes improved from 32 days to 26 days, thanks to the implementation of decentralized sequencing. This advancement not only speeds up reporting but also enhances the overall efficiency of the genomic surveillance system.
How did the transition to decentralized sequencing impact Listeria monocytogenes genomic surveillance in four Australian jurisdictions?
The transition to decentralized sequencing in four Australian jurisdictions initially caused mixed effects on turnaround times for Listeria monocytogenes genomic surveillance. While one jurisdiction seamlessly adapted to local sequencing, others experienced temporary delays. However, overall, this model increased system capacity and efficiency in the long run.
In what ways does whole-genome sequencing enhance the management of foodborne infections like listeriosis?
Whole-genome sequencing enhances the management of foodborne infections such as listeriosis by providing high-resolution characterization of pathogens. This enables health authorities to quickly identify outbreak clusters, link cases to food sources, and devise effective public health interventions, ultimately reducing the incidence of such infections.
What challenges were faced during the initial phase of decentralized sequencing for Listeria monocytogenes in Australia?
During the initial phase of decentralized sequencing for Listeria monocytogenes in Australia, some jurisdictions faced challenges, including increased turnaround times for genomic data reporting. These challenges underscored the need for well-defined sequence referral mechanisms to maintain efficiency and improve the system’s response to public health challenges.
Can the practices adopted in Australia’s decentralized sequencing model for Listeria monocytogenes be replicated in other countries?
Yes, the practices adopted in Australia’s decentralized sequencing model for Listeria monocytogenes can potentially be replicated in other countries. Establishing clear protocols for sequencing and data sharing can enhance genomic surveillance capacities worldwide, especially in managing foodborne diseases.
What statistical method was used to analyze the efficiency of the genomic surveillance system for Listeria monocytogenes?
The study analyzing the efficiency of the genomic surveillance system for Listeria monocytogenes employed the Kruskal-Wallis test to assess processing times across the years, offering insights into the improvements seen after the transition to decentralized sequencing.
| Key Point | Details |
|---|---|
| Turnaround Times | Median time from sample collection to genomic report decreased from 32 days in 2016 to 26 days in 2023. |
| Jurisdictional Sequencing | Four jurisdictions began sequencing samples locally, changing the dynamics of referral processes. |
| Impact of Decentralization | Decentralized sequencing improved the genomic surveillance system’s efficiency despite initial disruptions. |
| WGS Importance | Whole-genome sequencing is essential for effective outbreak investigations by linking clusters to sources. |
| Future Considerations | Australia’s experience could help other nations enhance their genomic surveillance capabilities. |
Summary
Decentralized Sequencing Listeria has significantly improved the genomic surveillance system in Australia, leading to faster and more efficient reporting of listeriosis cases. From 2016 to 2023, the median turnaround time for genomic surveillance reports dropped from 32 to 26 days. The transition to local sequencing in four jurisdictions has highlighted best practices and the importance of establishing clear referral mechanisms, contributing to a strengthened and responsive public health structure. Such advancements not only enhance Australia’s ability to manage public health threats but also serve as a valuable model for other countries aiming to improve their pathogen surveillance strategies.
The content provided on this blog (e.g., symptom descriptions, health tips, or general advice) is for informational purposes only and is not a substitute for professional medical advice, diagnosis, or treatment. Always seek the guidance of your physician or other qualified healthcare provider with any questions you may have regarding a medical condition. Never disregard professional medical advice or delay seeking it because of something you have read on this website. If you believe you may have a medical emergency, call your doctor or emergency services immediately. Reliance on any information provided by this blog is solely at your own risk.