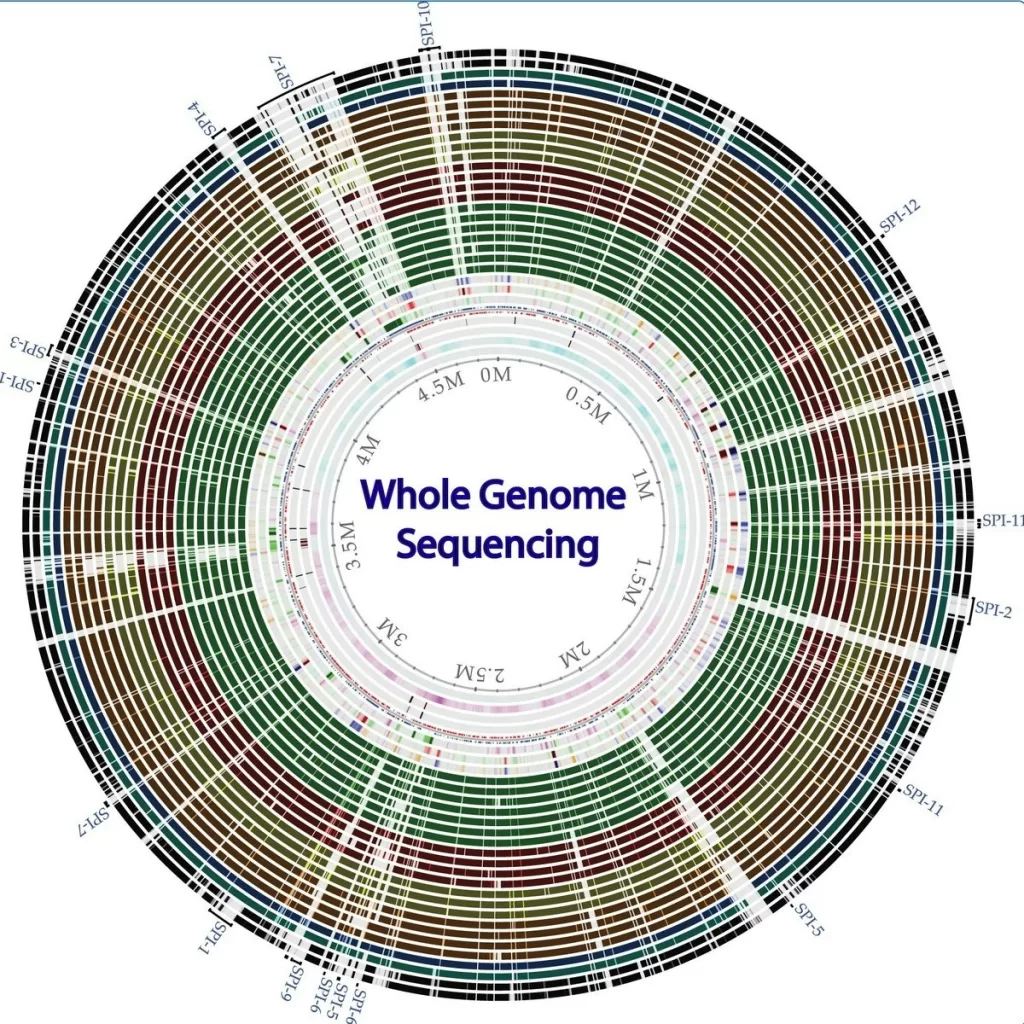
Whole-genome sequencing (WGS) has emerged as a groundbreaking method in microbiology, significantly enhancing our understanding of drug resistance in invasive pathogens such as *Streptococcus* spp. This advanced sequencing technology allows for comprehensive genetic analysis of bacterial isolates, paving the way for improved antimicrobial susceptibility testing. By leveraging bioinformatics in microbiology, researchers can now not only identify common resistance determinants but also streamline workflows through robust WGS frameworks. Notably, the Centers for Disease Control and Prevention’s Alaska Investigations Program has successfully utilized WGS to monitor *Streptococcus* drug resistance, improving public health responses. With a focus on invasive infections, the integration of WGS into routine practice is revolutionizing how we tackle infectious diseases across regions, particularly in Alaska where persistent challenges remain.
Genomic sequencing techniques, particularly whole-genome sequencing (WGS), represent a pivotal shift in how scientists and public health officials approach the detection of drug resistance in bacteria. The application of genomic analysis enables a detailed exploration of the genetic makeup of pathogens, particularly those causing invasive diseases such as those from the *Streptococcus* genus. Enhanced understanding of antimicrobial resistance mechanisms, achieved through bioinformatics and advanced analysis, plays a crucial role in effective antimicrobial susceptibility testing. This transformation in methodology offers a path toward better management of infectious diseases, leading to timely interventions and tailored treatment strategies. As demonstrated in recent research, adopting these next-generation sequencing workflows can significantly contribute to the battle against drug-resistant infections.
Understanding Whole-Genome Sequencing in Microbiology
Whole-genome sequencing (WGS) represents a revolutionary method in microbiological research, allowing for comprehensive insights into the genetic makeup of organisms, particularly pathogens. In the context of identifying drug resistance, WGS facilitates the determination of resistance genes and mechanisms by analyzing the entire genome of bacteria such as Streptococcus spp. This genomic approach significantly enhances our understanding of the evolutionary pathways these pathogens take, especially in regions like Alaska where distinct strains may emerge due to unique environmental factors.
Utilizing bioinformatics alongside WGS provides researchers with powerful tools to interpret the complex data generated. By mapping out the genetic information against known databases, scientists can quickly identify antimicrobial susceptibility and resistance profiles. This is particularly crucial for tracking the spread of drug-resistant Streptococcus strains, as it enables rapid public health responses to threats associated with invasive infections.
Frequently Asked Questions
What is whole-genome sequencing in microbiology?
Whole-genome sequencing (WGS) in microbiology refers to the comprehensive mapping of an organism’s entire DNA sequence. This technique is crucial for studying various bacteria, including those within the *Streptococcus* genus, as it allows for the identification of antimicrobial resistance genes and other genetic features important for understanding pathogenicity and drug resistance.
How is whole-genome sequencing used to study Streptococcus drug resistance?
Whole-genome sequencing is instrumental in identifying drug resistance in *Streptococcus* species. By analyzing the genetic makeup of isolated strains, researchers can pinpoint specific resistance determinants, such as ermTR and mef for macrolides, thereby enabling more accurate predictions of antimicrobial susceptibility and guiding effective treatment strategies.
What role does bioinformatics play in whole-genome sequencing?
Bioinformatics plays a critical role in whole-genome sequencing by providing the computational tools necessary for analyzing large volumes of genomic data. In the context of studying *Streptococcus* species, bioinformatics pipelines help interpret sequencing results, facilitating the understanding of genetic factors associated with drug resistance and aiding in the surveillance of infectious diseases.
What are the benefits of using a WGS workflow for antimicrobial susceptibility testing?
The WGS workflow offers numerous benefits for antimicrobial susceptibility testing, including increased accuracy and rapid data generation. By leveraging WGS, researchers can achieve a high concordance rate with traditional phenotypic testing methods, thus streamlining the process of monitoring and managing invasive *Streptococcus* infections.
How has WGS improved the study of infectious diseases in Alaska?
In Alaska, the transition to whole-genome sequencing has significantly enhanced the ability to monitor and respond to invasive infectious diseases. By employing WGS, the CDC’s Arctic Investigations Program can rapidly assess the antibiotic resistance profiles of *Streptococcus* species, leading to more effective public health interventions and improved patient outcomes.
What are some common resistance determinants found in Streptococcus species through WGS?
Common resistance determinants identified in *Streptococcus* species through whole-genome sequencing include ermB and ermTR for macrolides, tetM for tetracyclines, and mutations in gyrA and parC linked to levofloxacin resistance. Understanding these genetic markers is essential for effective antimicrobial susceptibility predictions.
Why might some researchers hesitate to adopt whole-genome sequencing workflows?
Some researchers may hesitate to adopt whole-genome sequencing workflows due to concerns about the accuracy of resistance predictions compared to traditional methods. However, studies have demonstrated that WGS can provide highly reliable predictions, often achieving over 99% concordance with phenotypic testing, which should alleviate such concerns.
How does the CDC Streptococcus Laboratory contribute to WGS in microbiology?
The CDC Streptococcus Laboratory plays a pivotal role in developing bioinformatics pipelines and workflows for whole-genome sequencing. These resources are vital for researchers studying *Streptococcus* species and contribute to improved accuracy in monitoring drug resistance and understanding the epidemiology of infections.
| Aspect | Details |
|---|---|
| Study Purpose | Evaluate WGS workflows for identifying drug resistance in Streptococcus spp. |
| Sample Collection | Isolates collected in Alaska from January 2019 to February 2021 (n=640) plus a historical subset. |
| Concordance Rates | 99.9% for GBS, 100% for GAS, and 99.98% for S. pneumoniae based on MIC predictions. |
| Common Resistance Genes | ermTR, ermB, mef for macrolides; tetM for tetracyclines; gyrA, parC for levofloxacin. |
| WGS Transition | Transitioned workflows to WGS in 2022 for faster monitoring of invasive diseases. |
| Methodology | DNA extraction, library preparation using Nextera Flex, and sequencing on MiSeq. |
| Overall Conclusion | WGS improves efficiency and accuracy of antimicrobial susceptibility predictions. |
Summary
Whole-genome sequencing (WGS) has emerged as a pivotal technique in modern microbiology, particularly for the identification of drug resistance in invasive Streptococcus species. The successful implementation of WGS at the Centers for Disease Control and Prevention’s Arctic Investigations Program demonstrates its ability to accurately predict antimicrobial susceptibility with high concordance rates. By transitioning from traditional phenotypic methods to WGS, healthcare professionals can achieve faster, more reliable data, ultimately enhancing the management of infectious diseases in challenging environments like Alaska.
The content provided on this blog (e.g., symptom descriptions, health tips, or general advice) is for informational purposes only and is not a substitute for professional medical advice, diagnosis, or treatment. Always seek the guidance of your physician or other qualified healthcare provider with any questions you may have regarding a medical condition. Never disregard professional medical advice or delay seeking it because of something you have read on this website. If you believe you may have a medical emergency, call your doctor or emergency services immediately. Reliance on any information provided by this blog is solely at your own risk.